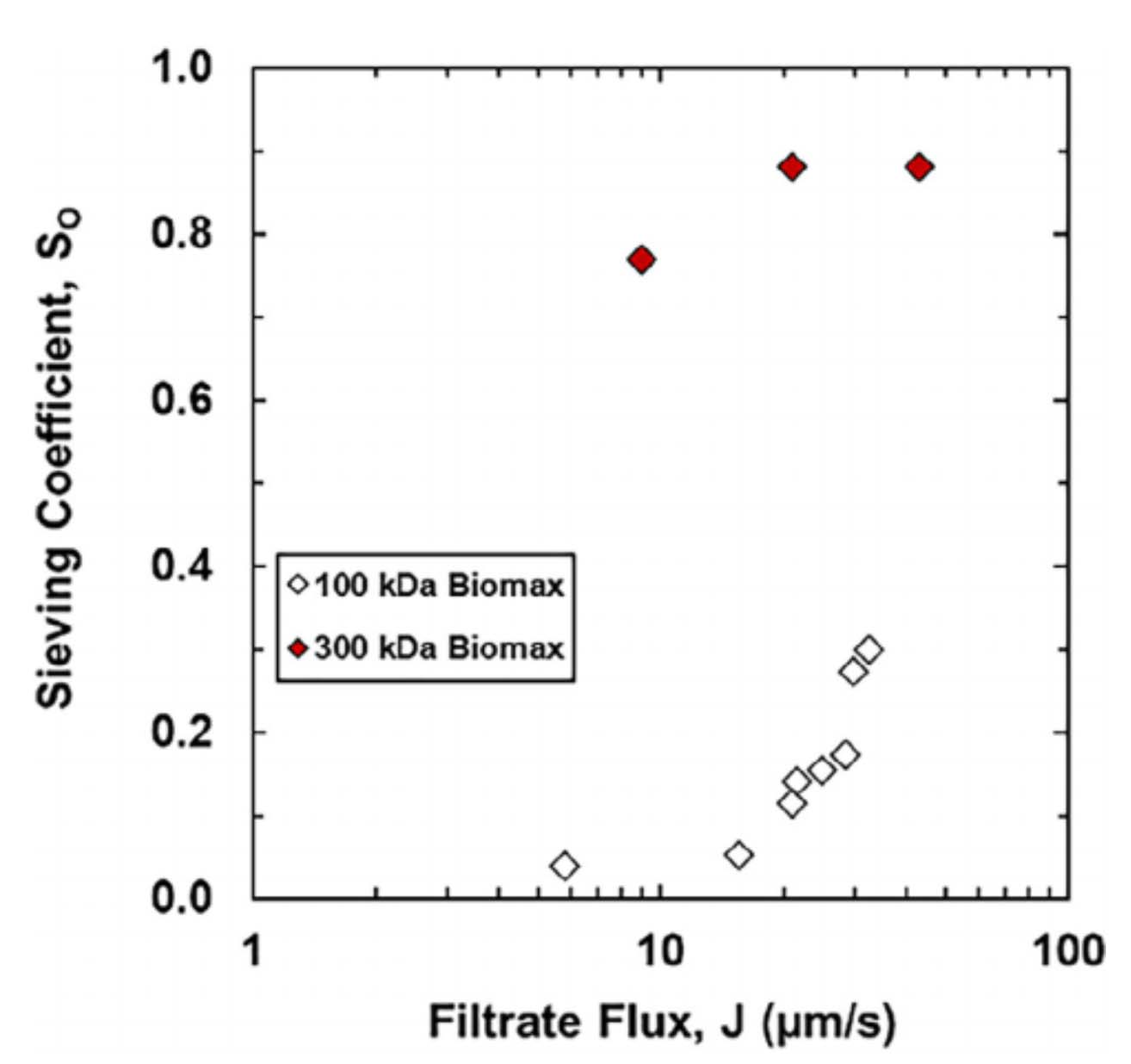
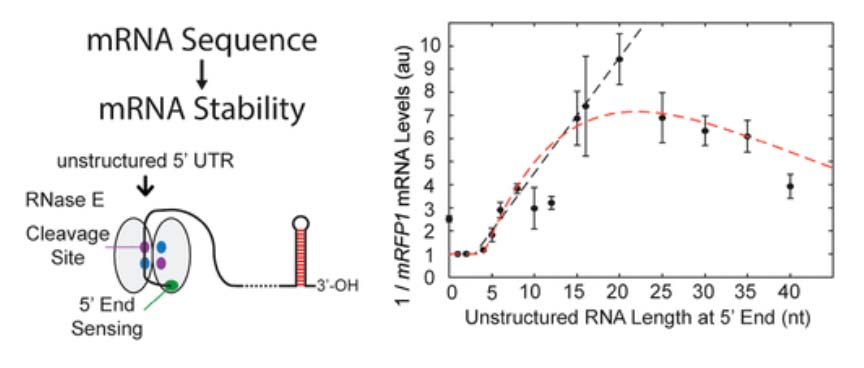
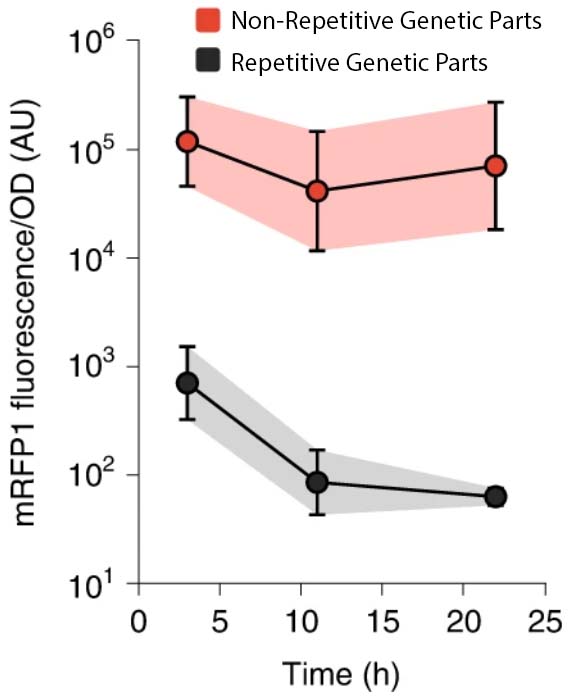
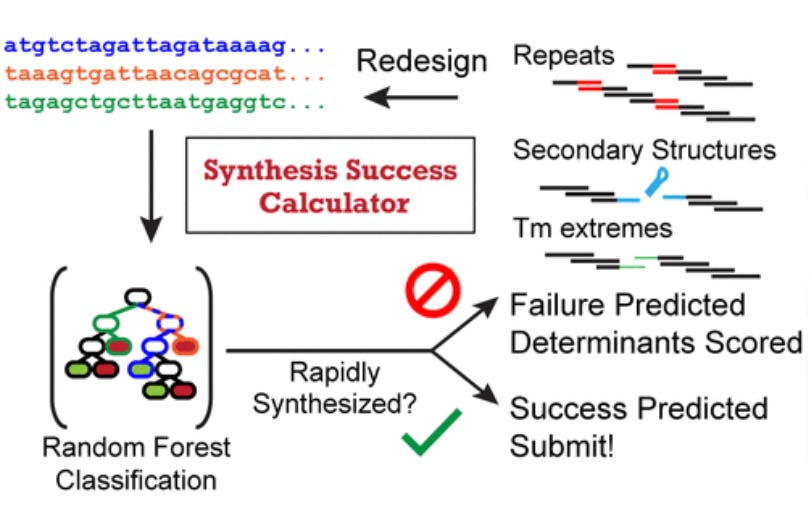
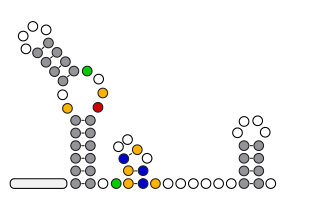The Biophysics of Gene Expression and Regulation
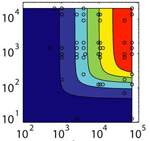
An organism's genes control its cellular fate: metabolism, self-replication, development, pathogenesis, and disease. Gene expression levels
are controlled by multiple layers of biomolecular interactions with its genetic template. With so many coupled interactions, it remains a challenge
to predict how DNA mutations affect gene expression levels.
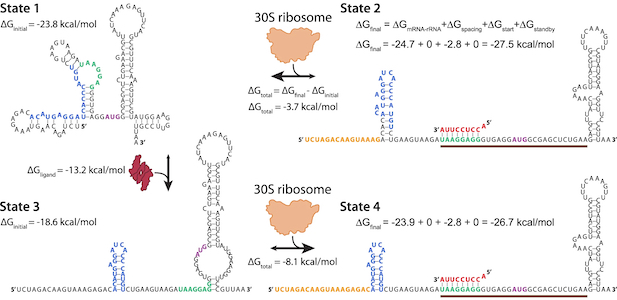
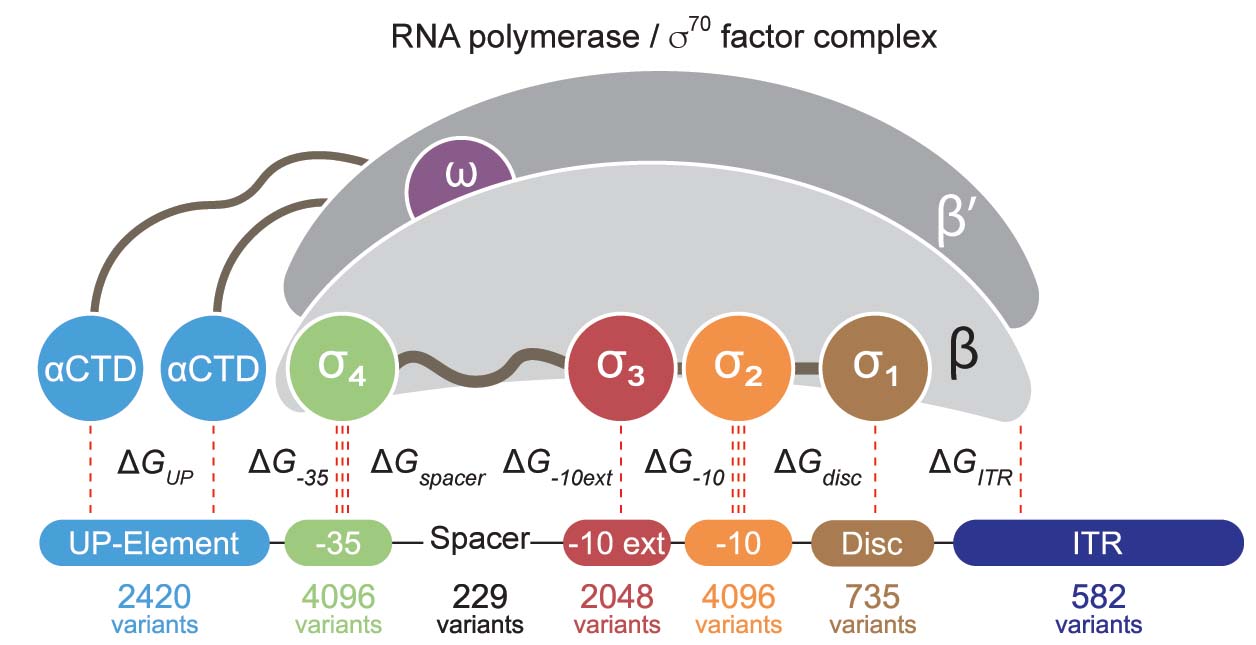
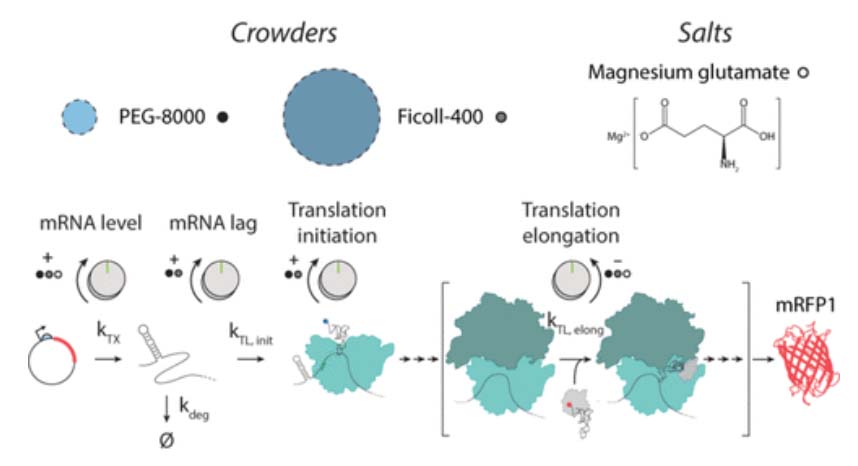
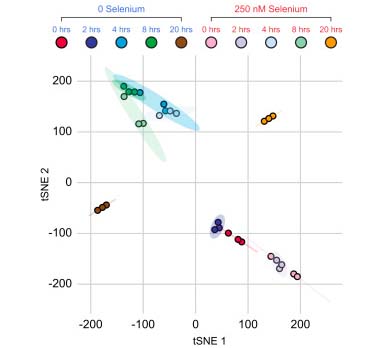
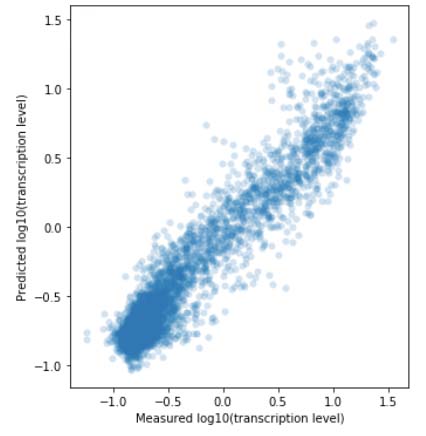
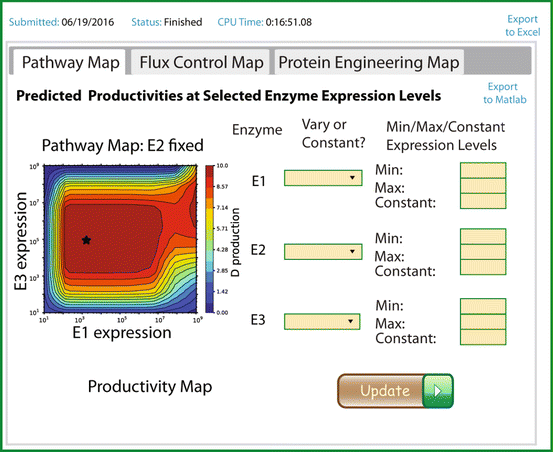
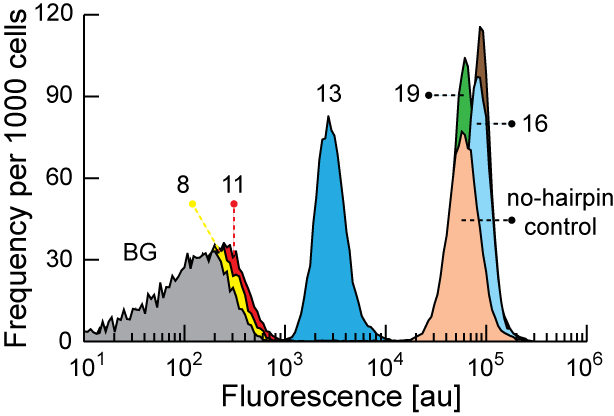
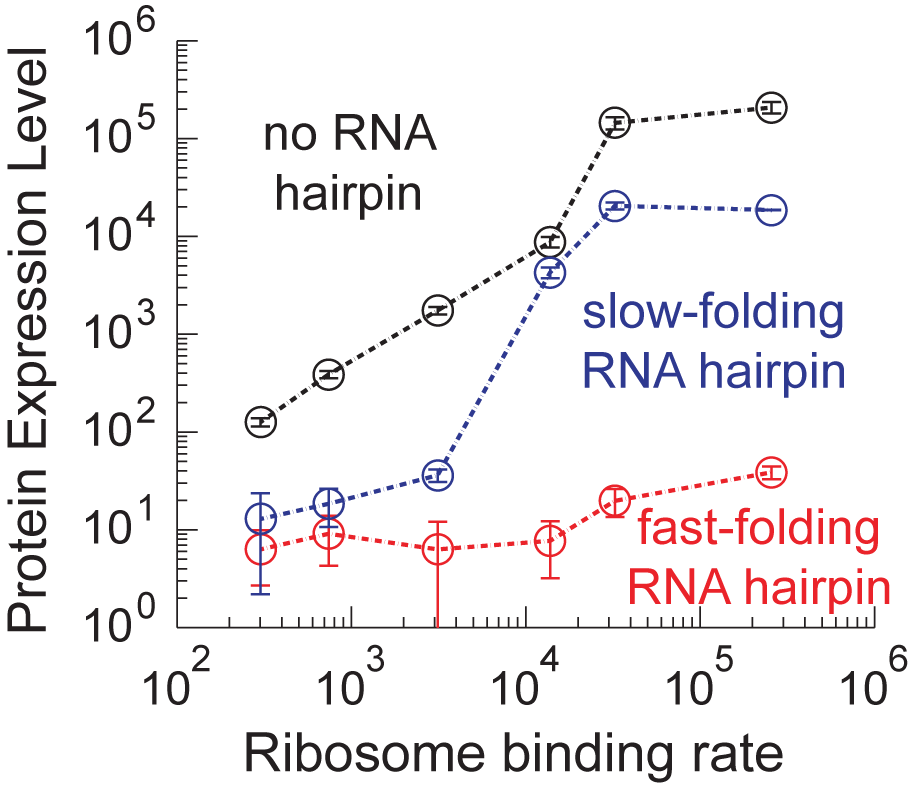
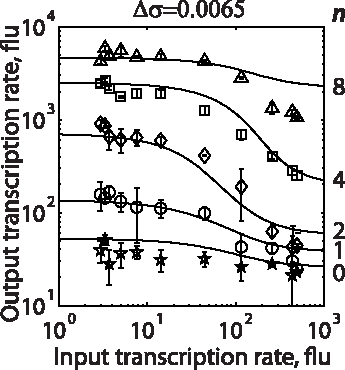
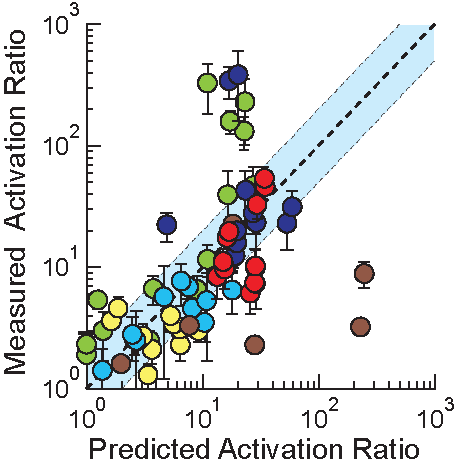
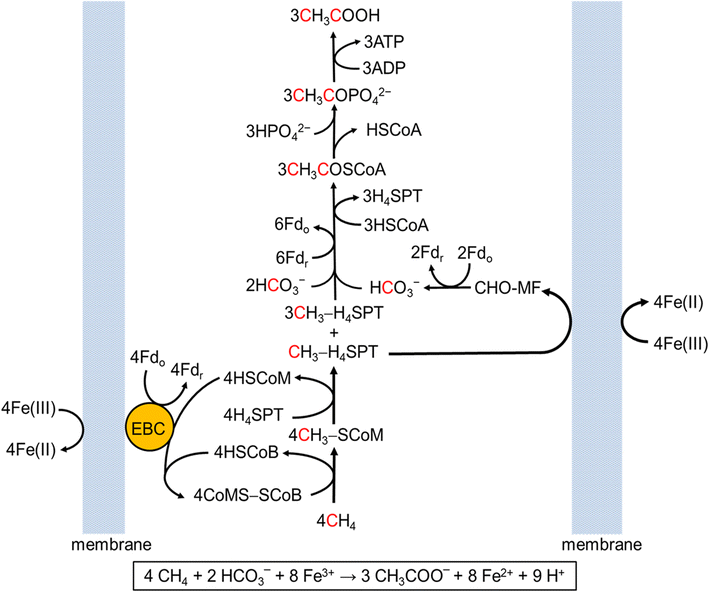
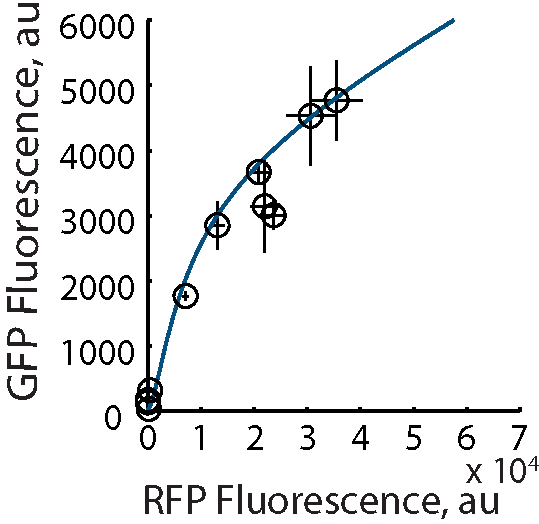
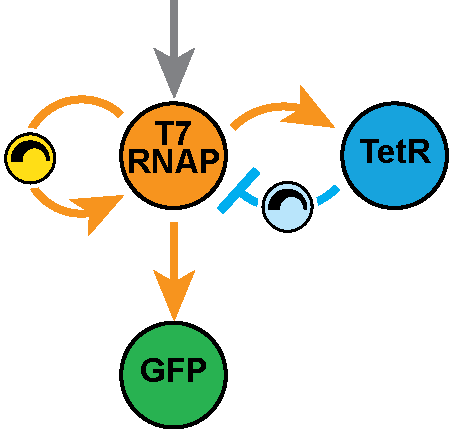
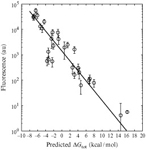
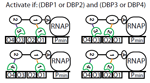
In the Salis Lab, we develop and experimentally validate biophysical models that predict
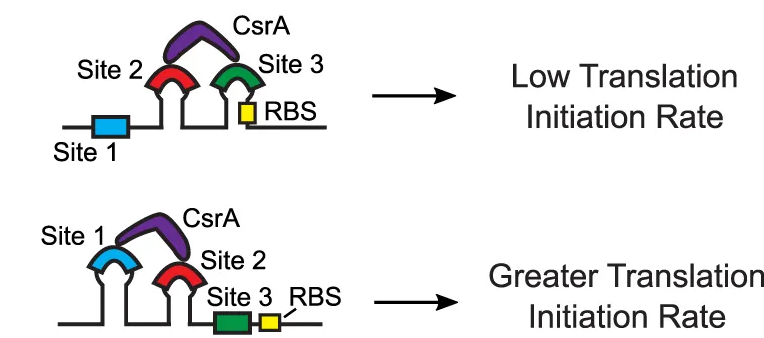
an organism's gene expression levels from its DNA sequence. Each model is highly reductionist, focusing on specific biomolecular interactions that control
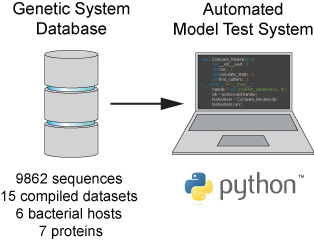
transcription rate, translation rate, or their regulation. We formulate low-parameter models using statistical thermodynamics and kinetics, parameterize them using rationally designed
series of experiments, and validate their predictions across hundreds of experiments using diverse DNA sequence sets. We then create user-friendly web interfaces so that the Life Science
community can use our biophysical models of gene expression for their own studies.
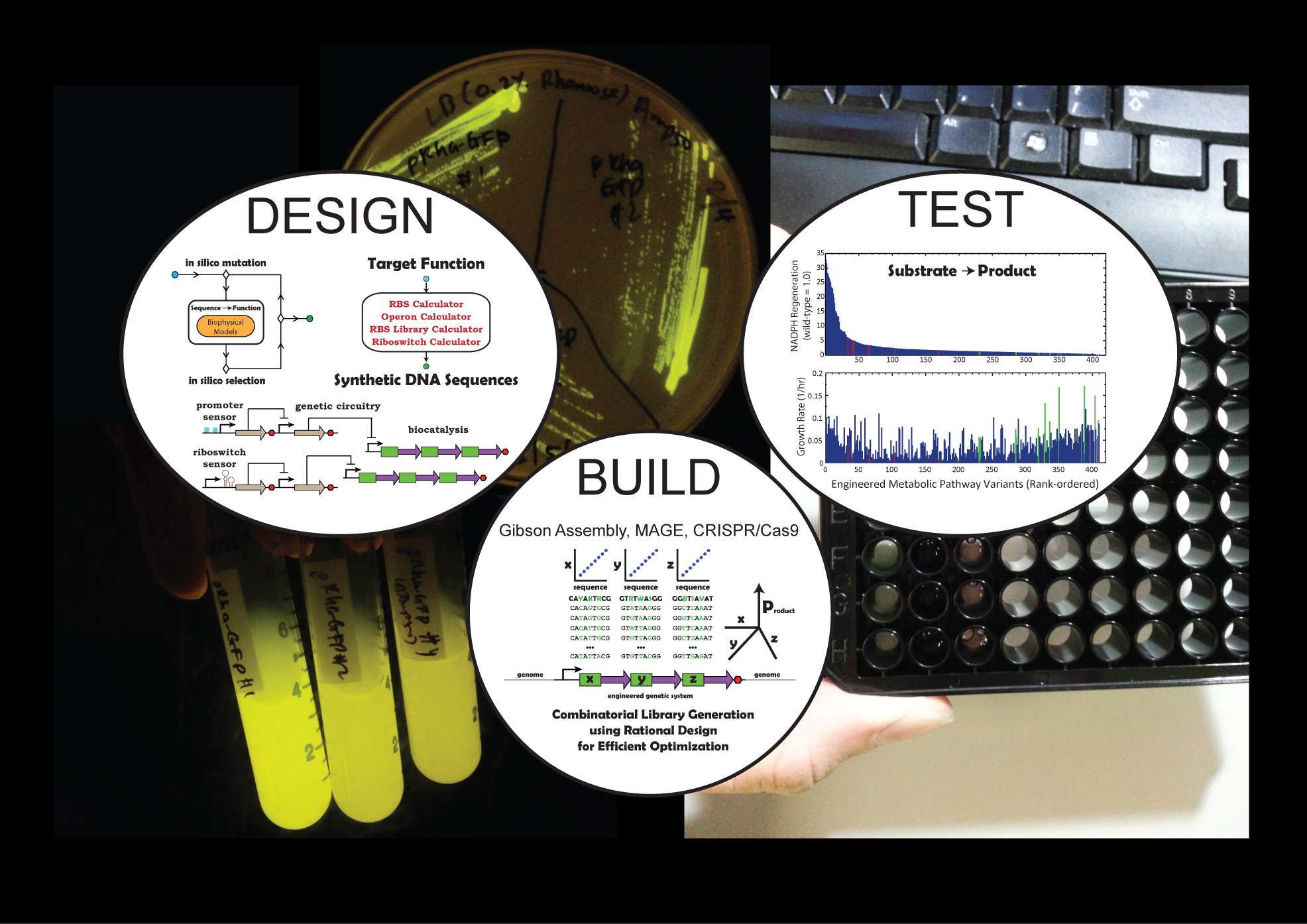 Welcome to the Salis Lab at UC RiversideWe develop predictive biophysical models and design algorithms
Welcome to the Salis Lab at UC RiversideWe develop predictive biophysical models and design algorithms Welcome to the Salis Lab at UC RiversideWe engineer bacteria, yeast, and algae to add new metabolic and sensing capabilities.
Welcome to the Salis Lab at UC RiversideWe engineer bacteria, yeast, and algae to add new metabolic and sensing capabilities.